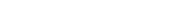| dc.contributor | Vall d'Hebron Barcelona Hospital Campus |
| dc.contributor.author | Piñana Moro, Maria |
| dc.contributor.author | Campos Martinez, Carolina |
| dc.contributor.author | Garcia Cehic, Damir |
| dc.contributor.author | Andrés Verges, Cristina |
| dc.contributor.author | Codina Grau, Gema |
| dc.contributor.author | Esperalba Esquerra, Juliana |
| dc.contributor.author | Sulleiro Igual, Elena |
| dc.contributor.author | Joseph Munné, Joan |
| dc.contributor.author | Saubí Roca, Narcís |
| dc.contributor.author | Colomer Castell, Sergi |
| dc.contributor.author | Martin Perez, Maria Carmen |
| dc.contributor.author | Castillo Oliveda, Carla |
| dc.contributor.author | Pumarola Suñé, Tomàs |
| dc.contributor.author | Rodríguez Frias, Francisco |
| dc.contributor.author | Antón Pagarolas, Andres |
| dc.contributor.author | Quer Sivila, Josep |
| dc.contributor.author | Rando Segura, Ariadna |
| dc.contributor.author | Gregori Font, Josep |
| dc.contributor.author | Esteban Mur, Juan Ignacio |
| dc.contributor.author | Cortese, Maria Francesca |
| dc.date.accessioned | 2022-04-22T13:19:54Z |
| dc.date.available | 2022-04-22T13:19:54Z |
| dc.date.issued | 2021-09-05 |
| dc.identifier.citation | Gregori J, Cortese MF, Piñana M, Campos C, Garcia-Cehic D, Andrés C, et al. Host-dependent editing of SARS-CoV-2 in COVID-19 patients. Emerg Microbes Infect. 2021;10(1):1777–89. |
| dc.identifier.issn | 2222-1751 |
| dc.identifier.uri | https://hdl.handle.net/11351/7379 |
| dc.description | SARS-CoV-2; Mutations; Quasispecies |
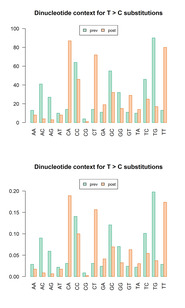
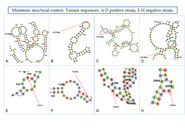
| dc.description.abstract | A common trait among RNA viruses is their high capability to acquire genetic variability due to viral and host mechanisms. Next-generation sequencing (NGS) analysis enables the deep study of the viral quasispecies in samples from infected individuals. In this study, the viral quasispecies complexity and single nucleotide polymorphisms of the SARS-CoV-2 spike gene of coronavirus disease 2019 (COVID-19) patients with mild or severe disease were investigated using next-generation sequencing (Illumina platform). SARS-CoV-2 spike variability was higher in patients with long-lasting infection. Most substitutions found were present at frequencies lower than 1%, and had an A → G or T → C pattern, consistent with variants caused by adenosine deaminase acting on RNA-1 (ADAR1). ADAR1 affected a small fraction of replicating genomes, but produced multiple, mainly non-synonymous mutations. ADAR1 editing during replication rather than the RNA-dependent RNA polymerase (nsp12) was the predominant mechanism generating SARS-CoV-2 genetic variability. However, the mutations produced are not fixed in the infected human population, suggesting that ADAR1 may have an antiviral role, whereas nsp12-induced mutations occurring in patients with high viremia and persistent infection are the main source of new SARS-CoV-2 variants. |
| dc.language.iso | eng |
| dc.publisher | Taylor & Francis |
| dc.relation.ispartofseries | Emerging Microbes and Infections;10(1) |
| dc.rights | Attribution 4.0 International |
| dc.rights.uri | http://creativecommons.org/licenses/by/4.0/ |
| dc.source | Scientia |
| dc.subject | COVID-19 (Malaltia) |
| dc.subject | Seqüència d'aminoàcids |
| dc.subject | Virologia |
| dc.subject.mesh | Coronavirus Infections |
| dc.subject.mesh | /virology |
| dc.subject.mesh | Amino Acid Sequence |
| dc.title | Host-dependent editing of SARS-CoV-2 in COVID-19 patients |
| dc.type | info:eu-repo/semantics/article |
| dc.identifier.doi | 10.1080/22221751.2021.1969868 |
| dc.subject.decs | infecciones por Coronavirus |
| dc.subject.decs | /virología |
| dc.subject.decs | secuencia de aminoácidos |
| dc.relation.publishversion | https://doi.org/10.1080/22221751.2021.1969868 |
| dc.type.version | info:eu-repo/semantics/publishedVersion |
| dc.audience | Professionals |
| dc.contributor.organismes | Institut Català de la Salut |
| dc.contributor.authoraffiliation | [Gregori J] Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Servei de Malalties Hepàtiques-Hepatitis Viral, Unitat del Fetge, Vall d’Hebron Hospital Universitari, Barcelona, Spain. Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd), Instituto de Salud Carlos III, Madrid, Spain. Roche Diagnostics SL, Barcelona, Spain. [Cortese MF, Saubí N] Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Servei de Bioquímica i Microbiologia, Vall d’Hebron Hospital Universitari, Barcelona, Spain. [Piñana M, Andrés C, Martin MC, Castillo C] Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Unitat de Virus Respiratoris, Servei de Microbiologia, Vall d’Hebron Hospital Universitari, Barcelona, Spain. [Campos C, Garcia-Cehic D, Quer J] Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Servei de Malalties Hepàtiques-Hepatitis Viral, Unitat del Fetge, Vall d’Hebron Hospital Universitari, Barcelona, Spain. Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd), Instituto de Salud Carlos III, Madrid, Spain. [Codina MG, Rando A, Esperalba J, Sulleiro E, Joseph J] Servei de Microbiologia, Vall d’Hebron Hospital Universitari, Barcelona, Spain. [Colomer-Castell S, Esteban JI] Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Servei de Malalties Hepàtiques-Hepatitis Viral, Unitat del Fetge, Vall d’Hebron Hospital Universitari, Barcelona, Spain. [Pumarola T] Servei de Microbiologia, Vall d’Hebron Hospital Universitari, Barcelona, Spain. Universitat Autònoma de Barcelona, Bellaterra, Spain. [Rodriguez-Frias F ] Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd), Instituto de Salud Carlos III, Madrid, Spain. Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Servei de Bioquímica i Microbiologia, Vall d’Hebron Hospital Universitari, Barcelona, Spain. Universitat Autònoma de Barcelona, Bellaterra, Spain. [Antón A] Vall d’Hebron Institut de Recerca (VHIR), Barcelona, Spain. Unitat de Virus Respiratoris, Servei de Microbiologia, Vall d’Hebron Hospital Universitari, Barcelona, Spain. Universitat Autònoma de Barcelona, Bellaterra, Spain |
| dc.identifier.pmid | 34402744 |
| dc.identifier.wos | 000754901600001 |
| dc.relation.projectid | info:eu-repo/grantAgreement/ES/PE2013-2016/RD16%2F0016%2F0003 |
| dc.relation.projectid | info:eu-repo/grantAgreement/ES/PE2017-2020/PI19%2F00301 |
| dc.rights.accessrights | info:eu-repo/semantics/openAccess |